WRF-Chem Tools for the Community
Please send all questions and discussions on these tools through the WRF-Chem Discussion Forum.
Announcements
- February 2022: Thanks to Caterina Mogno (University of Edinburgh, UK) the recently released EDGARv5.0 emission inventory is now available in MOZART speciation for use with anthro_emiss. See this link for more information and to download the data set.
- February 2022: A bug fix has been made to the epa_anthro_emis tool which corrects for an error in the mapping and specifically affected the results when mapping to higher resolutions. Thanks to a user for providing this fix.
- Update November 2021: Thanks to a WRF-Chem user, revised mozbc code for use with the new WRF hybrid coordinate is
available in the User Forum. - Please see the presentation on NCAR/ACOM Tools: "Biogenic, Fire, Lightning Emissions and Chemical Boundary Conditions" (August 2015).
- Please see the Best Practices 2015 Tutorial Presentation that includes example namelists for running with MOZCART and MOZART-MOSAIC.
- WRF-Chem Tutorial lectures are at the NOAA/ESRL WRF-Chem website.
Chemistry Options
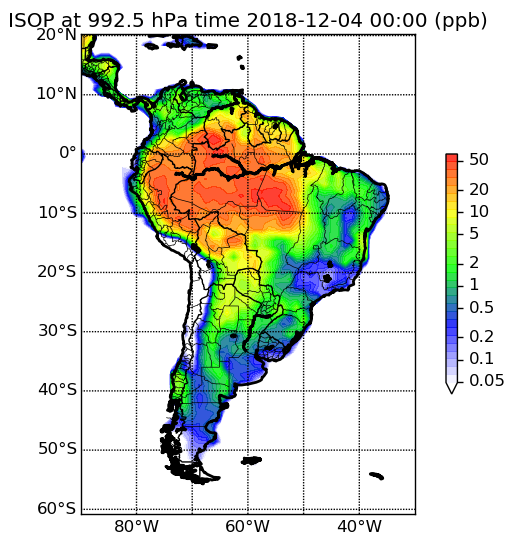
Isoprene emissions over South America, modeled with WACCM using the CAM-chem chemical mechanism. Click for larger image.
MOZART + GOCART (MOZCART) Chem_opt = 112 MOZART + MOSAIC 4 bins Chem_opt = 201 MOZART + MOSAIC 4 bins + AQCHEM Chem_opt = 202
The CAM-chem mechanism contains a detailed description of tropospheric inorganic chemistry and of organic species with 3 carbons or less, and a reasonably thorough treatment of isoprene chemistry. Larger alkanes and alkenes, terpenes, and aromatics are also included in the mechanism, albeit with lumping and simplification. The CAM-chem chemical mechanism and model description are published in Emmons et al. (2010). In MOZCART, the gas-phase chemistry is connected with the GOCART bulk aerosol scheme. More information on running WRF-Chem with the MOZCART chemical mechanism can be found in the MOZCART User's Guide. When using this option, please cite Pfister et al. (2011). Pre-processor tools for running WRF-Chem / MOZCART can be obtained from the Download section below.
In MOZART_MOSAIC, the gas-phase chemistry is coupled with the MOSAIC sectional aerosol scheme. The MOZART gas-phase chemistry was extended to include detailed treatment of monoterpenes (a-pinene, b-pinene, limonene) and MBO (Hodzic et al., in prep.), aromatics, HONO, C2H2, as well as an updated isoprene oxidation scheme (Knote et al., 2014). More information on running WRF-Chem with the MOZART/MOSAIC chemical mechanism can be found in the MOZART_MOSAIC User's Guide.
With WRF-Chem V3.9 a new TUV photolysis option has been included (phot_opt=4). This option requires an additional data file. Please refer to the Instructions on how to run WRF-Chem with the new TUV photolysis option.
Preprocessors
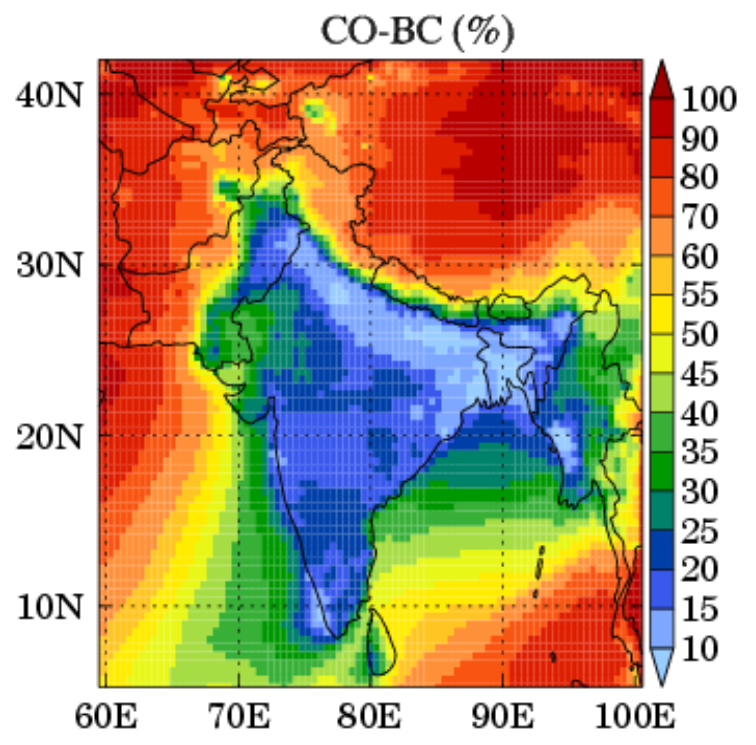
Surface spatial distributions of average total CO mixing ratios (ppbv) and relative contribution of CO‐BC (%) to total CO during January–February 2008. Figure 7 of Kumar, R., M. Naja, G. G. Pfister, M. C. Barth, and G. P. Brasseur (2013), Source attribution of carbon monoxide in India and surrounding regions during wintertime, J. Geophys. Res. Atmos., 118, 1981–1995, doi: 10.1002/jgrd.50134. Click for larger image.
We ask users of the WRF-Chem preprocessor tools to include in any publications the following acknowledgement:
"We acknowledge use of the WRF-Chem preprocessor tool {mozbc, fire_emiss, etc.} provided by the Atmospheric Chemistry Observations and Modeling Lab (ACOM) of NCAR."
mozbc: Creates lateral boundary and initial conditions from global chemistry model output.
NCAR/ACOM has developed a program to create time-varying chemical lateral boundary conditions for WRF-Chem from CAM-chem output. For questions about running mozbc please use our WRF-Chem Discussion Forum contacting wrf-chem-mozbc.
For modifications required to run mozbc with the new WRF hybrid coordinate, please see the discussions in the User Forum (Subject line: [WRF-Chem mozbc] mozbc in WRF 3.9 or Greater)
To obtain mozbc, see the Download section below.
IMPORTANT: Please note that mozbc is not yet working with the hybrid sigma-pressure vertical coordinate introduced with WRF V3.9.
To obtain CAM-chem output files use the CAM-chem Download page. Note that mozbc also has the option to set species to a single fixed value. This is especially relevant for long-lived and well distributed species (e.g. CH4, H2 or N2O) if they are not on the global model output.
The reference for the CAM-chem output is:
Tilmes, S., Emmons, L.K., Buchholz, R.R. & The CESM2 Development Team, (2022). CESM2.2/CAM-chem Output for Boundary Conditions. UCAR/NCAR - Atmospheric Chemistry Observations and Modeling Laboratory. Subset used† XXX, Accessed* dd mmm yyyy, DOI: https://doi.org/10.5065/XS0R-QE86.
†Please fill in the "subset used" with region and/or date of the subset you used (e.g. Lat: -10 to 10, Lon: 100 to 150, September 2015 - February 2016).
*Please fill in the "Accessed" date with the day, month, and year that you last accessed the data (e.g. - 5 Aug 2011).
Near-real-time global chemical forecast output from the WACCM model is also available for use as regional model boundary conditions.
Mapping of WACCM or CAM-chem gas-phase and aerosol species to WRF-Chem chemistry is provided in this document: CESM-WRFchem_aerosols_20190822.pdf.
Upper boundary conditions for chemical species
Download input files for running WRF-Chem V3.3.1 with Chemical Upper Boundary Conditions: UBC_inputs.tar Presentation describing upper boundary condition: WRF_workshop_UBC.pdf
bio_emiss: Create MEGAN input for WRF-Chem.
To obtain bio_emiss, see the Download section below.
anthro_emiss: Create anthropogenic emissions files from global emission inventories
This is a fortran based preprocessor to create WRF-Chem ready anthropogenic emissions files (wrfchemi__ or wrfchemi_{00z,12z}_) from global inventories on a lat/lon basis. Users are strongly advised to consult the README files before compiling and using the code. See the Download section below.
anthro_emiss with EDGAR-HTAP emissions: For users who like to use the anthro_emiss tool with the global EDGAR-HTAP emission inventory (http://edgar.jrc.ec.europa.eu/national_reported_data/htap.php) we provide inputs needed for mapping to the MOZART-MOSAIC and MOZART-GOCART chemical options. This package has been provided by Rajesh Kumar (rkumar@ucar.edu).
The recently released EDGARv5.0 emission inventory is now available in MOZART speciation for use with anthro_emiss. See this link for more information and to download the data set (provided by Caterina Mogno, University of Edinburgh, UK. Email).
NEW: EPA_ANTHRO_EMIS: This tool allows users to create WRF-Chem compatible hourly anthropogenic emission input files from Sparse Matrix Operator Kernel (SMOKE) Modeling System netcdf output. Please consult the EPA_ANTHRO_EMIS User Guide and the README included in the download before using the tool. A sample input data set containing the U.S. EPA 2014v2 emissions is available for download: EPA 2014v2 Input Data.
For use with the U.S. EPA 2014v2 emissions, users also will need to download the sectorlist_2014fd_nata file and place it into the data directory.
U.S.EPA NEI 2017 input data are now available for download. These data have been made available to the community by Alison Eyth and Barron Henderson at U.S. EPA. For more information on NEI 2017 see the U.S. EPA report “Derived Estimates of Air Quality for 2017”. EPA_ANTHRO_EMIS can be used with the new data set but it will require updates to the namelist input file. See the README in the download for more information. Click to download an example input file for the MOZCART-T1 mechanism.

Aerial view of the Rabbit Foot Fire in Idaho on 13 Aug 2018. Smoke from the Rabbit Foot Fire was sampled by the NSF/NCAR C-130 during three separate WE-CAN research flights and several times for longer durations by ground-based mobile labs. (Photo credit: Rebecca Hornbrook) Click for larger image.
fire_emiss: Create fire emissions files
Fortran based preprocessor for creating fire emission inputs for WRF-Chem when running with plumerise and also for creating fire emission inputs for the CAM-Chem global models. The fire emissions inventory is based on the Fire Inventory from NCAR (FINN). Both software (fire_emis.tgz) and required FINN input data sets are available at the FINN download page.
The fire_emis.tgz file when uncompressed {tar -zxf fire_emis.tgz} yields three directories {data_files, src, and test} and two readme files {README.WRF.fire and README.GLB.fire }. The data_files directory is empty and is where users should put the FINN files and the wrfinput_dfile(s). The test directory contains two test namelist input files, one for creating WRF inputs and another for creating global inputs. Users are highly advised to read the README files before using the fire emission utility.
Aircraft Emissions Preprocessor
This is an IDL based preprocessor to create WRF-Chem ready aircraft emissions files (wrfchemaircraft_) from a global inventory in netcdf format. Please consult the README file for how to use the preprocessor. The emissions inventory is not included, so the user must provide their own. Download the preprocessor files.
Post-Processors
Integrated Reaction Rates (IRR)
The MOZART chemistry options (chem_opt = 112, 201, 203) can now be run with a new diagnostic that outputs the time integrated gas-phase reaction rates (IRR) of the chemical mechanism. This capability is similar to the integrated reaction rate analysis in the Community Multiscale Air Quality Modeling System (CMAQ). (October 2017)
Download
Use the Processors Download page to register and retrieve the above software packages.
FINN fire emissions are available at the FINN download page.
CAM-chem output files can be downloaded from here.